
In-Fusion cloning is a very versatile method that can be used to create seamless gene fusions.Ĭapture PCR products by TA or GC cloning.Ĭhoose traditional TA cloning or high-efficiency GC cloning.įor convenience, the common TA cloning vector and Lucigen's GC cloning vector are provided.Īnnealing two oligonucleotides forms a double-stranded product. Simulate Clontech's In-Fusion clone by inserting up to eight fragments in the vector. The linearized vector can be generated by enzymatic digestion or inverse PCR. Select the fragment to be fused and its direction, and SnapGene will design primers. Gibson Assembly is a popular seamless cloning method. Simulate Gibson Assembly by fusing up to eight fragments or by inserting up to eight fragments into the vector. Select the fragments to be assembled, and SnapGene will design primers.

The color of history highlights abrupt changes.Īnalog gateway? BP clone or LR clone, or both at the same time.įor convenience, common donor vectors and destination vectors are provided. Select the mutagenic primer and press the button to view the modified plasmid. Use mutagenic primers for site-directed mutagenesis. Select the fragment to be ligated and its direction, and SnapGene will design primers. The product file stores templates and primers in its history.įusion fragments by overlap extension PCR Use your own primers, or ask SnapGene to design primers automatically. Visualize all aspects of the cloning process. Use tool tips to avoid problematic restricted sites. SnapGene will automatically mark the site with an asterisk. Show unique restriction sites in bold, or select the automatically defined Unique Cutters or Unique 6+ Cutters enzyme group.ĭon't be fooled because the restriction site is blocked by Dam, Dcm or EcoKI methylation. Select a part of a DNA or protein sequence and annotate features using flexible GenBank compatible controls.Ĭonfirm that the restricted website is suitable for cloning. Other functions of your choice can be added to the custom database. Use SnapGene's extensive database to find common features in DNA sequences. The color is visible in both the Map and Sequence view.Īnnotate frequently used functions automatically, or annotate new functions manually. Set the selected DNA or amino acid sequence to one of ten colors.Ĭolor two DNA strands or protein sequences. When copying and pasting the sequence, the function is automatically transferred.Įdit the ends of linear DNA sequences to add or remove overhangs or phosphates. The ancestral protein or DNA sequence can be reproduced as a separate file. View the automatically generated protein sequence graph history. The checkbox toggles between compact or fully expanded display. Sort the annotated feature list by name, location, size, color or type. The same analysis can be performed on selected parts of the protein. View protein molecular weight, extinction coefficient, isoelectric point and amino acid composition. Use 1- or 3-letter amino acid codes with related characteristics to view protein sequences. View multiple views of protein sequences.Ĭustomize the display of areas, sites, keys and sequence colors. Use the multi-function controls to adjust the zoom factor and display area. Use the proprietary MICA algorithm to find the sequence in the chromosome immediately. Use SnapGene's efficient data processing function to scan large DNA sequences with thousands of annotation functions. Ancestor sequence can be restored as a separate file

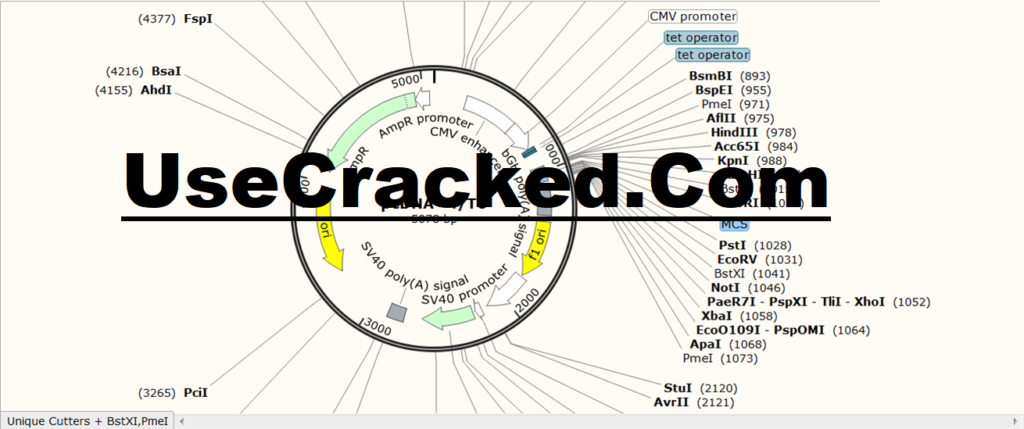
View the automatically generated graphical history of DNA constructs. Sort the hybrid primer list by name, length, color, binding site, directionality or melting temperature. Sort the annotated feature list by name, location, size, color, directionality or type. Cut sites can be displayed as numbers or rows, sorted by name or frequency.
#Snapgene viewer change color series#
Single chain mode displays a compact overview with color features.Ĭhoose from a series of enzyme groups. Use double-stranded mode to view enzyme sites, with features of translation, primers, and DNA color. The map can be in circular or linear format. Customize the display of enzyme sites, features, primers, ORF, DNA color, etc.


 0 kommentar(er)
0 kommentar(er)
